backbones filtering
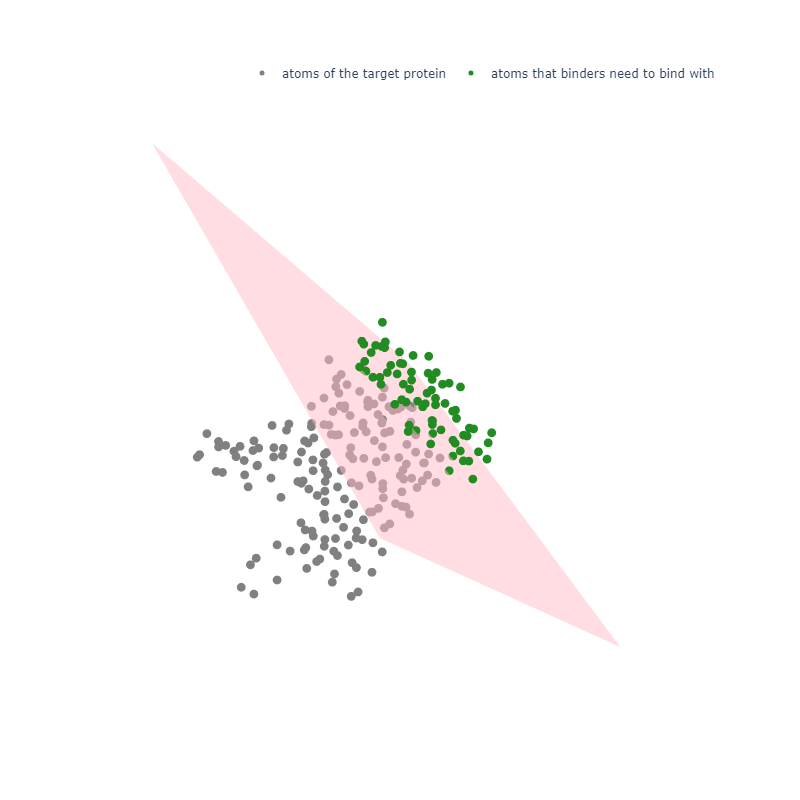
In design binders via RFdiffusion and ProteinMPNN-FastRelax, at the beginning of sequence design and binder assessment script mpnn_af.slm, we could perform a fitering step to remove useless backnones.
In my opinion, this is a very heloful step, because it greatly increase the speed and efficiency of desigining reasonable binders. For example, in my target.pdb, I only expect designed binders could bind with the outside part of the target.pdb (see fig.1).