large-language model (LLM)
Recently, I read some papers, and I found one commonality that they treat protein data as text data. The natural language processing (NLP) methods seem highly transferable to the computational biology. For example, the Tansformer model is specially effective for language understanding, such as the language translation. ESM3 smartly used the mechanisms of Transformer in its own model and specially for the protein task.
A consensus is developing that underlying these sequences is a fundamental language of protein biology that can be understood using language models (6–11)
I would like to keep sharing what I am learning.
- Hayes, Tomas, et al. "Simulating 500 million years of evolution with a language model." bioRxiv (2024): 2024-07.
Simulating 500 million years of evolution with a language model
article
In Introduction section, authors state why they started this work. Firstly, architectures and representations within language models, especially transformer-based ones, map very well onto the world of protein. Secondly, "Great strength produces miracles". Increasing scale means increasing accuracy.
A number of language models of protein sequences have now been developed and evaluated (5–10, 12–17). It has been found that the representations that emerge within language models reflect the biological structure and function of proteins (6–8, 18), and are learned without any supervision on those properties (19, 20), improving with scale (5, 21). In the field of artificial intelligence, scaling laws have been found that predict the growth in capabilities with increasing scale, describing a frontier in compute, parameters, and data.
So, they give the ESM3 model, a multimodal generative language model (sequence, strcuture, function) . It was trained on 2.78 billion proteins, 771 billion unique tokens, and has 98 billion parameters.
After the ESM3 is trained (training process is supervised), they set up an input sequence, and asked the ESM3 model to generate a group of protein who are much likely to achieve in lab experiments.
We set out to create a functional green fluorescent protein (GFP) with low sequence similarity to existing ones. We chose the functionality of fluorescence because it is difficult to achieve, easy to measure, and one of the most beautiful mechanisms in nature.
They then performed a first experiment with 88 designs.
Among the generations that we synthesized, we found a bright fluorescent protein at far distance (58% identity) from known fluorescent proteins. Similarly distant natural fluorescent proteins are separated by over five hundred million years of evolution
The SEM3, a multi-modal transformer-based generative model, makes it true to find or design new proteins based on certain conditions.
We have found that language models can reach a design space of proteins that is distant from the space explored by natural evolution, and generate functional proteins that would take evolution hundreds of millions of years to discover. Protein language models do not explicitly work within the physical constraints of evolution, but instead can implicitly construct a model of the multitude of potential paths evolution could have followed
notes
What excits me a lot in this article? (I am thinking these points might be used in my tasks.)
- the way of integrating sequence, structure and function together.
- the application of transformer model.
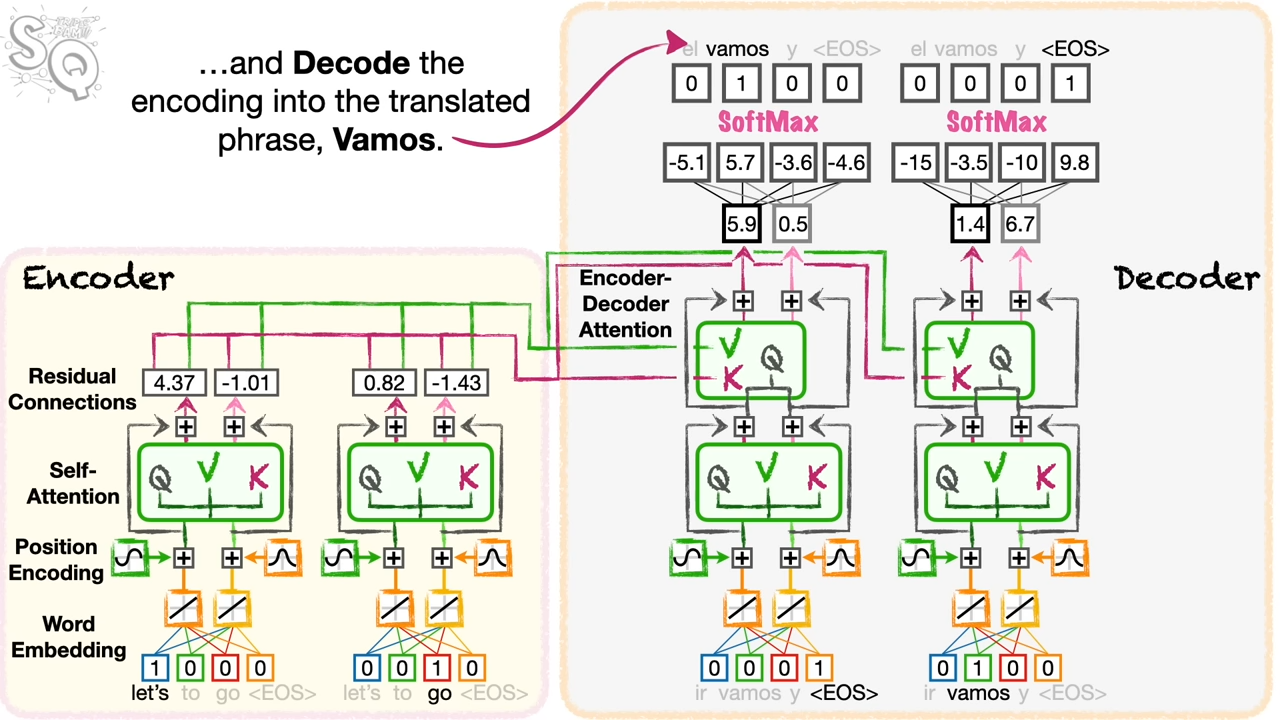
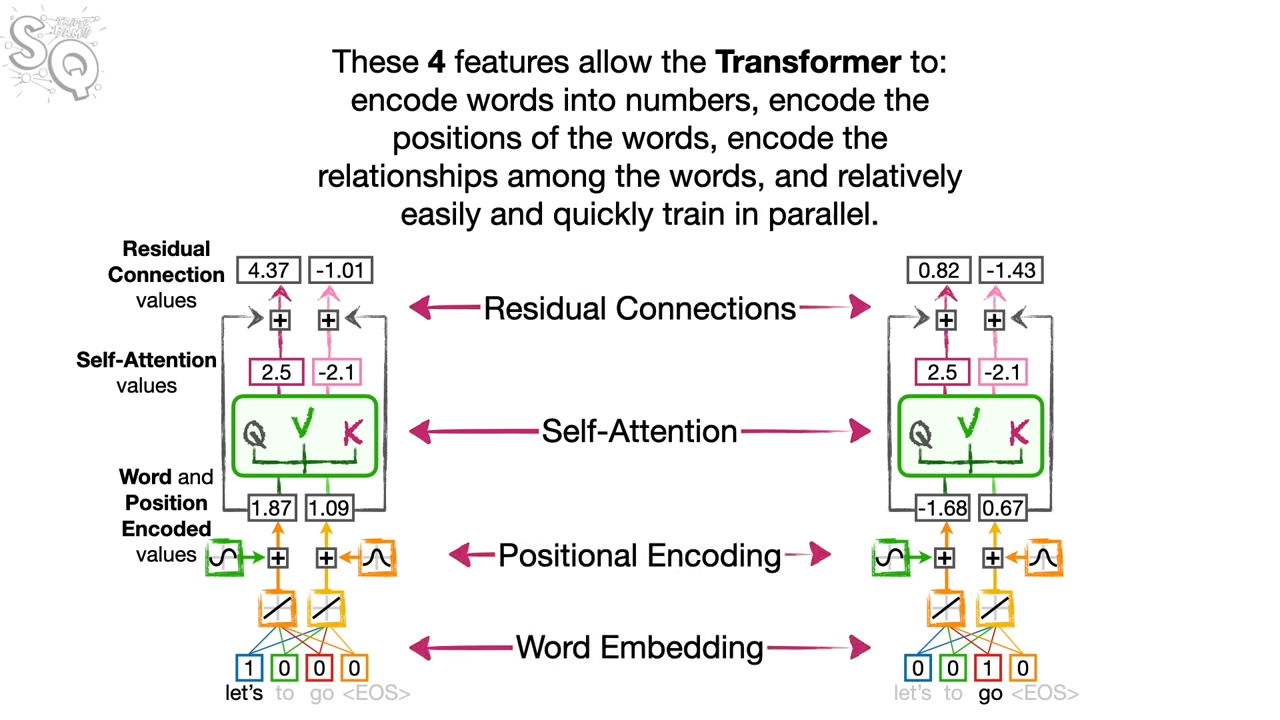
I spend some time on the Transformer model. In the following example, it is a translation from lets go to vamos.